This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
General Information
Protein Name: Solute carrier family 2, facilitated glucose transporter member 4 (GLUT4)
Classification: Belongs to the major facilitator superfamily, Sugar transporter (TC 2.A.1.1) family, Glucose transporter subfamily.
Subcellular Location: Primarily in the perinuclear region. Undergoes continuous recycling to the plasma membrane and rapid reinternalization. The dileucine internalization motif is critical for intracellular sequestration [1].
Protein Name: Solute carrier family 2, facilitated glucose transporter member 4 (GLUT4)
Classification: Belongs to the major facilitator superfamily, Sugar transporter (TC 2.A.1.1) family, Glucose transporter subfamily.
Subcellular Location: Primarily in the perinuclear region. Undergoes continuous recycling to the plasma membrane and rapid reinternalization. The dileucine internalization motif is critical for intracellular sequestration [1].
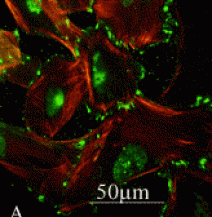
Visceral epithelial cells. Green: GLUT4 immunofluorescence. Red: actin cytoskeleton staining. Retrieved from www.medscape.com/viewarticle/516533_3
Tissue Specificity: Skeletal and cardiac muscles; brown and white fat.
Sequence and Motifs
Isoforms: There is only one known isoform of GLUT4
NCBI Reference Sequence: NP_001033.1
Length: 509 AA
Sequence Diversity: Polymorphism
Sequence and Motifs
Isoforms: There is only one known isoform of GLUT4
NCBI Reference Sequence: NP_001033.1
Length: 509 AA
Sequence Diversity: Polymorphism
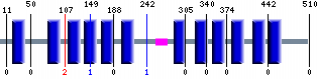
SMART Motif Search
Prosite Motif Search
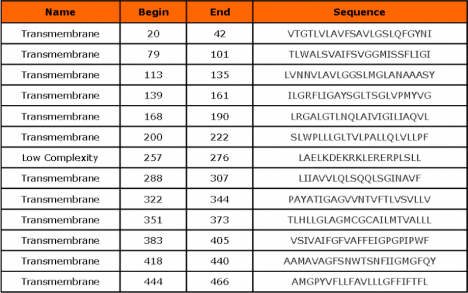
SUGAR_TRANSPORT_1, PS00216; Sugar transport proteins signature 1 (PATTERN)
Site: 144 - 169: LiGAYsGLtsglvpmYvgEiapthlR
Sequences known to belong to this class detected by the pattern: the majority of transporters with 23 exceptions Other sequence(s) detected in Swiss-Prot: 53.
SUGAR_TRANSPORT_2, PS00217; Sugar transport proteins signature 2 (PATTERN)
Site: 340 - 356: SVLLVERAGRRtlhll.G
Sequences known to belong to this class detected by the pattern: the majority of transporters with 20 exceptions Other sequence(s) detected in Swiss-Prot: 67.
Site: 144 - 169: LiGAYsGLtsglvpmYvgEiapthlR
Sequences known to belong to this class detected by the pattern: the majority of transporters with 23 exceptions Other sequence(s) detected in Swiss-Prot: 53.
SUGAR_TRANSPORT_2, PS00217; Sugar transport proteins signature 2 (PATTERN)
Site: 340 - 356: SVLLVERAGRRtlhll.G
Sequences known to belong to this class detected by the pattern: the majority of transporters with 20 exceptions Other sequence(s) detected in Swiss-Prot: 67.
UniPort
When we searched GLUT4 in UniPort, we found two more domains: 1) The SRFBP1-binding domain from AA 7-13 and [3] 2) the internalization dileucine domain from AA 489-490 [1].
Also we found in UniPort that the residue Asparagin at position 57 is glycosilated [4]. The Serine at position 488 is also phosphorylated.
When we searched GLUT4 in UniPort, we found two more domains: 1) The SRFBP1-binding domain from AA 7-13 and [3] 2) the internalization dileucine domain from AA 489-490 [1].
Also we found in UniPort that the residue Asparagin at position 57 is glycosilated [4]. The Serine at position 488 is also phosphorylated.
Phylogeny
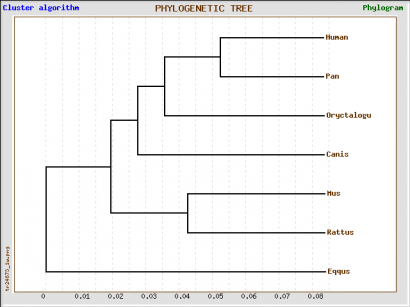
Phylogeny.fr tree
Phylogeny.fr tree
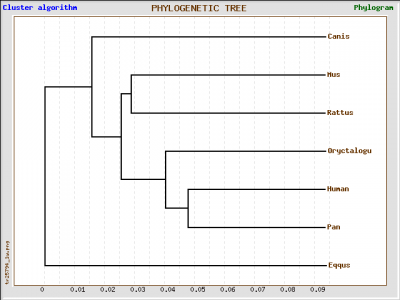
GenBee Tree from ClustalW Alignment
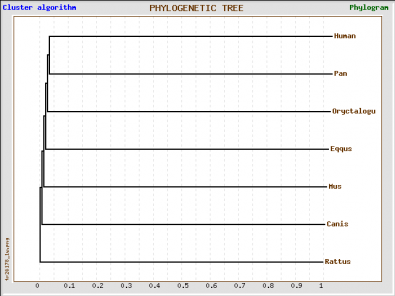
GenBee Tree from Muscle Alignment
GenBee Tree from T-Coffee Alignment
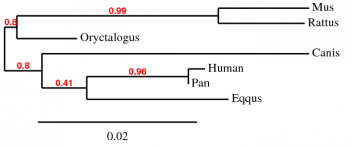
We obtained four different trees, one using Phylogeny.fr and the other three using GenBee. For Phylogeny.fr we just pasted the amino acid sequence, and then the software used MUSCLE to align the sequence and GBlocks algorithms to curate the tree. For GenBee, we had to paste an alignment of the sequences we wanted to compare. The three different alignments were obtained using ClustalW, MUSCLE, and T-COFFEE. They gave different trees because the alignments use different penalties for every mismatch.
The four trees show significant differences. First, the phylogeny.fr tree puts the horse GLUT4 as the most similar protein to human after the one from chimpanzee, whereas in the Muscle and ClustalW alignments they are the furthest away. The trees from the ClustalW and T-Coffee alignments seem to be less accurate becuase they place the rabbit (a rodent) closer to primates than to other rodents such as rat and mouse.
Since it has ethical implications to use chimpanzees as model organisms, and it is impractical to use big mammals, the best candidates for further studies would be rabbit, mouse and rat. According to the Muscle alignment tree, these three animals have the closest protein sequence to human among the species shown.
The four trees show significant differences. First, the phylogeny.fr tree puts the horse GLUT4 as the most similar protein to human after the one from chimpanzee, whereas in the Muscle and ClustalW alignments they are the furthest away. The trees from the ClustalW and T-Coffee alignments seem to be less accurate becuase they place the rabbit (a rodent) closer to primates than to other rodents such as rat and mouse.
Since it has ethical implications to use chimpanzees as model organisms, and it is impractical to use big mammals, the best candidates for further studies would be rabbit, mouse and rat. According to the Muscle alignment tree, these three animals have the closest protein sequence to human among the species shown.
Protein Interactors
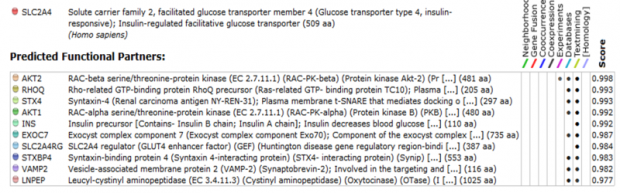
STRING
STRING
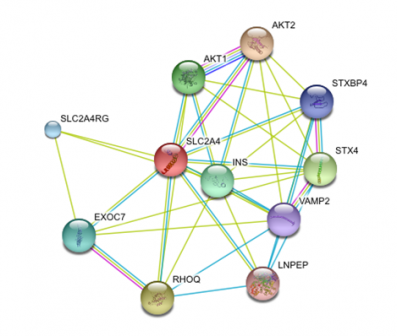
In order to find the functional partners of Glut4, we used STRING. STRING is a piece of software that searches databases for known and predicted interactions, including direct (physical) and indirect (functional) interactions. An advantage of this software is that it recognizes homologs from other species and predicts an interaction based on conservation.
When we run a search for Glut4, we found that most of the proteins associated with it are proteins related to membrane trafficking and vesicle fusion. An extremely useful feature from string is that it gives you the chance to follow the evidence of each interaction found. In that way we were able to see the gene onthologies of the interactors.
When we run a search for Glut4, we found that most of the proteins associated with it are proteins related to membrane trafficking and vesicle fusion. An extremely useful feature from string is that it gives you the chance to follow the evidence of each interaction found. In that way we were able to see the gene onthologies of the interactors.
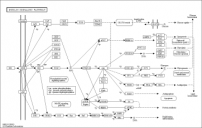
KEGG PATHWAY
Using KEGG PATHWAY we found three pathways in which Glut4 is involved. This pathways match with the gene onthologies found for Glut4.
Using KEGG PATHWAY we found three pathways in which Glut4 is involved. This pathways match with the gene onthologies found for Glut4.
References
- Verhey, K.J., Birnbaum, M.J. (1994) A Leu-Leu sequence is essential for COOH-terminal targeting signal of GLUT4 glucose transporter in fibroblasts. J. Biol. Chem. 269:2353-2356(1994)
- Kusari, J., Verma, U.S., Buse, J.B., Henry R.R., Olefsky J.M. (1991). Analysis of the gene sequences of the insulin receptor and the insulin-sensitive glucose transporter (GLUT-4) in patients with common-type non-insulin-dependent diabetes mellitus. J. Clin. Invest. 88:1323-1330
- Lalioti, V.S., Vergarajauregui, S., Pulido, D., Sandoval, I.V.(2002). The insulin-sensitive glucose transporter, GLUT4, interacts physically with Daxx. Two proteins with capacity to bind Ubc9 and conjugated to SUMO1. J. Biol. Chem. 277:19783-19791
- Wollscheid, B., Bausch-Fluck, D., Henderson, C., O'Brien, R., Bibel, M., Schiess, R., Aebersold, R., Watts, J.D. (2009). Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27:378-386
Carlos Gil del Alcazar, [email protected], last updated on 3/28/10