This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Further Directions
Type 2 diabetes is a complex disorder, meaning that not just one gene affects it. Inherited susceptibility towards Type 2 diabetes (and several other diseases) is associated with multiple single nucleotide polymorphisms. This means that SNP's confer an individual more propensity to developing a disease. Virtually polymorphisms on any gene directly or indirectly associated with the glucose level sensing, insulin signaling/production pathways, vesicle membrane trafficking, glucose transport, etc, might be involved in the susceptibility towards diabetes Type 2. In order to identify such polymorphisms genome-wide scans of affected people have to be performed. And then association analysis has to be performed in order to confirm the role of such polymorphisms in diabetes causation.
V383I
From our protein analysis of Glut4, we found that a polymorphism was identified in which a Valine (383V) is replaced by an Isoleucine residue. Such polymorphism has only been found in patients with Type 2 diabetes [1], however, not all patients affected with diabetes type 2 have this polymorphism. So we believe that only a subpopulation of the patients have this polymorphism. First we checked whether V383 is conserved among other species (Figure 1), and we found that it is indeed conserved. This conservation shows its importance in proper Glut4 function.
Type 2 diabetes is a complex disorder, meaning that not just one gene affects it. Inherited susceptibility towards Type 2 diabetes (and several other diseases) is associated with multiple single nucleotide polymorphisms. This means that SNP's confer an individual more propensity to developing a disease. Virtually polymorphisms on any gene directly or indirectly associated with the glucose level sensing, insulin signaling/production pathways, vesicle membrane trafficking, glucose transport, etc, might be involved in the susceptibility towards diabetes Type 2. In order to identify such polymorphisms genome-wide scans of affected people have to be performed. And then association analysis has to be performed in order to confirm the role of such polymorphisms in diabetes causation.
V383I
From our protein analysis of Glut4, we found that a polymorphism was identified in which a Valine (383V) is replaced by an Isoleucine residue. Such polymorphism has only been found in patients with Type 2 diabetes [1], however, not all patients affected with diabetes type 2 have this polymorphism. So we believe that only a subpopulation of the patients have this polymorphism. First we checked whether V383 is conserved among other species (Figure 1), and we found that it is indeed conserved. This conservation shows its importance in proper Glut4 function.
Experiments
Yeast-2-Hybrid
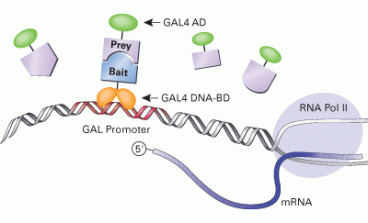
It is possible that this polymorphism is somehow affecting the way Glut4 interacts with other proteins. This might affect the way Glut4 is regulated. In order to find whether Glut4 has unknown binding partners that interact specifically with the region where the polymorphism is, we want to perform a yeast-2-hybrid experiment. First we want to use the following system to find the proteins that interact with Glut4 naturally:
In which the Exon 9 of Glut4 (bait) is fused to the GAL4 DNA-binding domain, and a comprehensive library (prey) of unknown proteins is fused to the GAL4-activating domain. If GAL4 AD and GAL4 DNA-BD bind indirectly (through the bait and prey interaction) GAL will be transcribed and thus expressed. Then it is very easy to detect the colonies expressing GAL by a color change in the medium due to a reaction catalyzed by GAL. Such colonies' DNA is sequenced and the interacting protein is identified. So if we find interacting partners to Exon 9, we would repeat the experiment using the same system but this time using the mutant version of Exon 9.
We might find that the Exon9 of Glut4 naturally interacts with proteins with onthologies such as membrane trafficking regulation or vesicle fusion. However, with the V383I version we might find another set of interactors or no interactors at all. This would interfere with the proper translocation of Glut4 to the membrane.
Mass Spectrometry
There has not been a significant study in posttranslational (PTM) modifications in Glut4. PTM's are very important in proper folding and regulation of proteins. For instance, kinases activate other proteins by phosphorylating them providing key steps in regulation. For this reason we want to do a correlation study between the PTM state of Glut4 and Diabetes in patients displaying the disease.
In order to do this, we want to run a mass spec from protein extracts of muscle or fat tissue from both healthy subjects and patients with Type 2 diabetes. The controls (samples from healthy subjects) will tell us whether there are PTM's other than the ones previously found. The samples from the patients with Type 2 diabetes, will tell us whether there are missing PTM's (compared to the controls).
So the first step would be to prepare the sample. Since we know the sequence, we can easily calculate both the molecular weight and the isoelectric point of Glut4. Using a 2-D gel, then we can roughly isolate the protein to simplify the sample for mass spec. We want to simplify the sample as much as possible because there is a big similarity between Glut4 and other glucose transporters, this might mislead the results of mass spec (similar peaks would be obtained for any glucose transporter). After that, we would digest the sample with trypsin and run the sample through a tandem mass spec. This will give us a series of peaks to which a software will assign a score which is a statistic of how confident we are of that specific fragment being part of Glut4. Also the difference of mass between the peaks will be able to tell us weather a residue has a PTM on it.
We expect to find PTM's in the controls that are absent in the samples from diabetic patients. Such result would support the hypothesis that missing PTM's in Glut4 results in the inability of Glut4 to be translocated. We also expect that not all the patients will be missing such PTM's since diabetes can occur because of other irregularities in any other gene related to glucose intake.
We might find that the Exon9 of Glut4 naturally interacts with proteins with onthologies such as membrane trafficking regulation or vesicle fusion. However, with the V383I version we might find another set of interactors or no interactors at all. This would interfere with the proper translocation of Glut4 to the membrane.
Mass Spectrometry
There has not been a significant study in posttranslational (PTM) modifications in Glut4. PTM's are very important in proper folding and regulation of proteins. For instance, kinases activate other proteins by phosphorylating them providing key steps in regulation. For this reason we want to do a correlation study between the PTM state of Glut4 and Diabetes in patients displaying the disease.
In order to do this, we want to run a mass spec from protein extracts of muscle or fat tissue from both healthy subjects and patients with Type 2 diabetes. The controls (samples from healthy subjects) will tell us whether there are PTM's other than the ones previously found. The samples from the patients with Type 2 diabetes, will tell us whether there are missing PTM's (compared to the controls).
So the first step would be to prepare the sample. Since we know the sequence, we can easily calculate both the molecular weight and the isoelectric point of Glut4. Using a 2-D gel, then we can roughly isolate the protein to simplify the sample for mass spec. We want to simplify the sample as much as possible because there is a big similarity between Glut4 and other glucose transporters, this might mislead the results of mass spec (similar peaks would be obtained for any glucose transporter). After that, we would digest the sample with trypsin and run the sample through a tandem mass spec. This will give us a series of peaks to which a software will assign a score which is a statistic of how confident we are of that specific fragment being part of Glut4. Also the difference of mass between the peaks will be able to tell us weather a residue has a PTM on it.
We expect to find PTM's in the controls that are absent in the samples from diabetic patients. Such result would support the hypothesis that missing PTM's in Glut4 results in the inability of Glut4 to be translocated. We also expect that not all the patients will be missing such PTM's since diabetes can occur because of other irregularities in any other gene related to glucose intake.
References
1. Kusari, J., Verma, U.S., Buse, J.B., Henry R.R., Olefsky J.M. (1991). Analysis of the gene sequences of the insulin receptor and the insulin-sensitive glucose transporter (GLUT-4) in patients with common-type non-insulin-dependent diabetes mellitus. J. Clin. Invest. 88:1323-1330
1. Kusari, J., Verma, U.S., Buse, J.B., Henry R.R., Olefsky J.M. (1991). Analysis of the gene sequences of the insulin receptor and the insulin-sensitive glucose transporter (GLUT-4) in patients with common-type non-insulin-dependent diabetes mellitus. J. Clin. Invest. 88:1323-1330
Carlos Gil del Alcazar, [email protected], last updated on 5/11/10